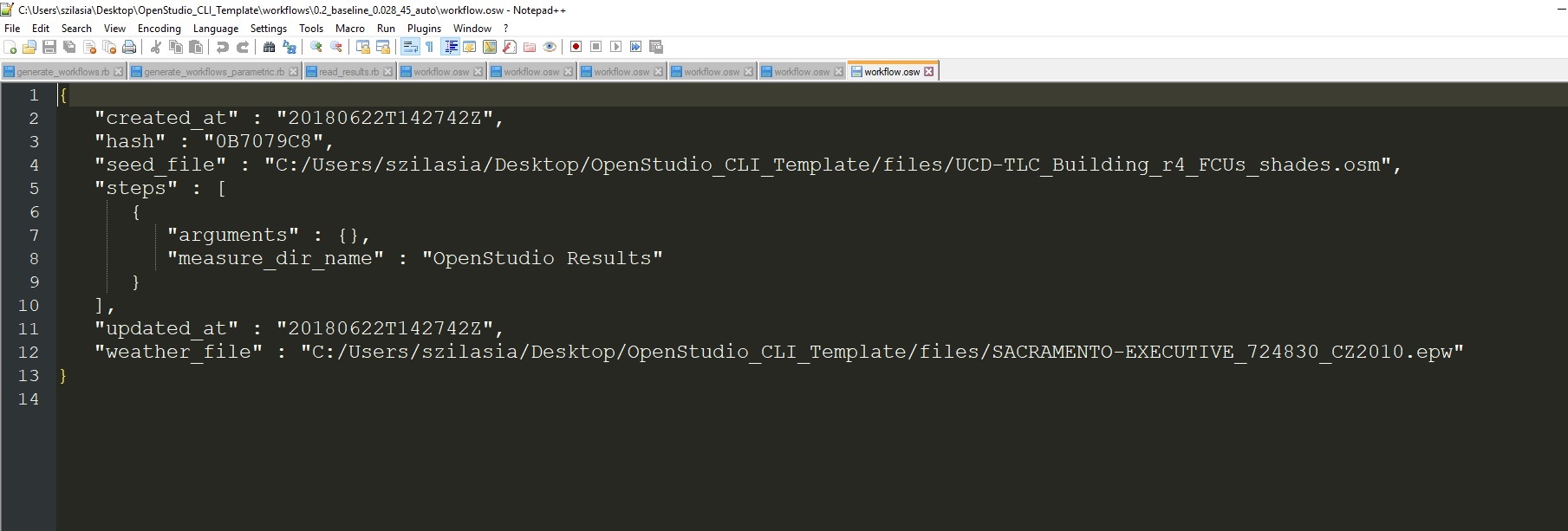
One model, one osw.
Documentation for the osw structure found here and schema here.
You can build an osw programmatically using the WorkflowJSON object
As an example, this is what your script might look like:
# the logic in the parametric script is built around making an .osw file
def make_osw(run_name, osm_filepath, epw_filepath, model_measure_steps = [], energyplus_measure_steps = [], reporting_measure_steps = [])
puts "Making directory #{run_name}"
FileUtils.mkdir "workflows/#{run_name}"
osw_filepath = OpenStudio::Path.new("#{Dir.pwd}/workflows/#{run_name}/workflow.osw")
osw = OpenStudio::WorkflowJSON.new
osw.setSeedFile(osm_filepath)
osw.setWeatherFile(epw_filepath)
if !model_measure_steps.empty?
measure_type = OpenStudio::MeasureType.new("ModelMeasure")
osw.setMeasureSteps(measure_type, model_measure_steps)
end
if !energyplus_measure_steps.empty?
measure_type = OpenStudio::MeasureType.new("EnergyPlusMeasure")
osw.setMeasureSteps(measure_type, energyplus_measure_steps)
end
if !reporting_measure_steps.empty?
measure_type = OpenStudio::MeasureType.new("ReportingMeasure")
osw.setMeasureSteps(measure_type, reporting_measure_steps)
end
osw.saveAs(osw_filepath)
puts "#{run_name} osw written"
return true
end
# define measures to add to the model
measure1 = OpenStudio::MeasureStep.new("measure1")
measure1.setArgument("arg1","some string argument value")
measure2 = OpenStudio::MeasureStep.new("measure2")
measure2.setArgument("arg1",0.5)
# path to an epw file and a list of paths to the multiple seed models
# use ruby Dir.glob or related script to pull all .osms paths in a directory into an array
# e.g. osm_files = Dir.glob(File.join("seed_models/**","*.osm"))
epw_path = "path/to/epw/tmy3.epw"
model_paths = ["/path/to/file/seed1.osm","/path/to/file/seed2.osm","/path/to/file/seed3.osm"]
# convert them to OpenStudio::Path objects
epw_path = OpenStudio::Path.new(epw_path)
model_paths = model_paths.map {|x| OpenStudio::Path.new(x)}
model_paths.each do |path|
run_name = path.to_s.split("/")[-1].chomp(".osm") + "_measure1and2"
makeOSW(run_name, path, epw_path, [measure1, measure2], [], [])
end
Alternatively, you could adjust the measure so instead of making a directory for each .osw, it names each .osw and dumps them all in the same directory
 Inside the first folder
Inside the first folder
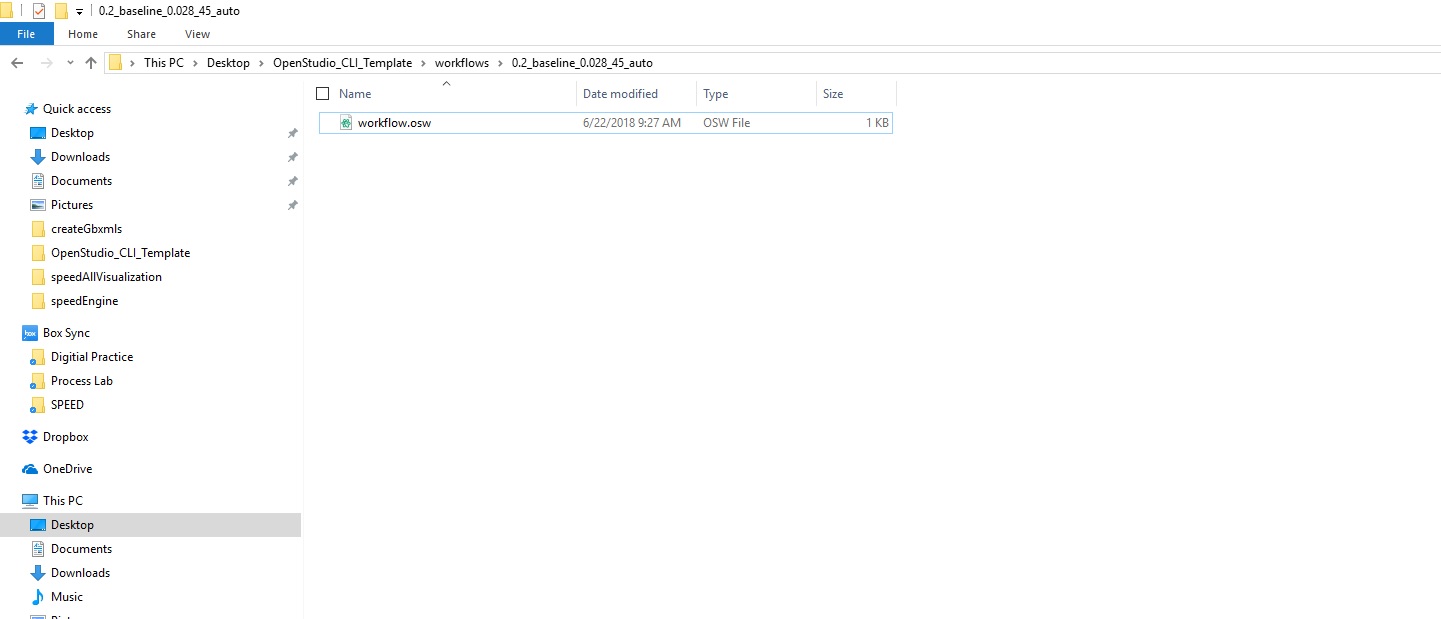 The OSW inside the first folder - representing a workflow for a seed model
The OSW inside the first folder - representing a workflow for a seed model